Annotations¶
mzmine uses a variety of annotation methods. These methods include spectral library matches, compound database annotations, lipid annotations, and manual annotation.
Spectral library matches¶
To annotate compounds by matching experimental MS2 (or MSn) spectra against spectral libraries using the Spectral library search module. mzmine supports several open library formats (see here) and also features a workflow to build your own spectral libraries.
Compound database annotations¶
The compound database annotation matches experimental MS1 data (m/z, RT, CCS, RI) against a library using the Local compound database search module. This library may be downloaded or generated in-house.
Lipid annotations¶
mzmine's lipid annotations are a rule-based fragmentation framework to confidently annotate lipids based on MS2 and/or MS1 data using the Lipid annotation module. You can also define custom lipids and create your own annotation rules.
Manual annotation¶
To annotate compounds manually use the the context menu in the feature table (right click on a row) and select "Identification" → "Annotate manually". The result is stored as a compound database match.
Preferred annotation¶
The preferred annotation is a combination of spectral library matches, compound database annotations, and lipid annotations. The most confident annotation is chosen according to a set of community guidelines. mzmine supports ranking of annotations based on the MSI minimum reporting standards, the Exposomics annotation levels (Celma et. al., Schymanski et. al.) and default mzmine sorting.
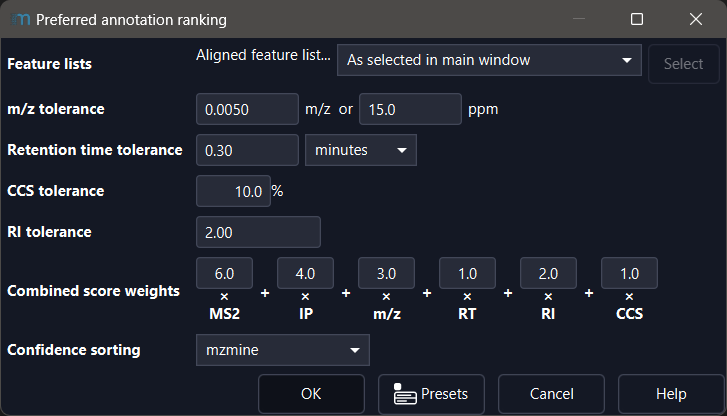
Preferred annotation ranking (module)¶
The preferred annotation ranking module allows you to modify the scoring tolerances and scoring
weights of the annotation quality summary (AQS) and the confidence
sorting for the preferred annotations.
Parameters¶
Tolerance parameters¶
The m/z, RT, CCS, and RI tolerances describe the maximum permissible tolerance to achieve a match and thus a non-zero score. Larger deviations will be indicated by an orange label in the AQS.
Combined score weights¶
The weights define how important an individual score is to the combined score.
Confidence sorting¶
Defines how the preferred annotation is determined and how annotations are sorted in the feature table when sorting by AQS. The available options are mzmine, Exposomics, and MSI and are described below. After the sorting by confidence scale determines the main level, the annotations are sorted internally by the combined sore.
Default annotation ranking in mzmine¶
The default sorting algorithm in mzmine is adapted from the Exposomics levels, but specifically weights a lipid annotation with diagnostic fragments higher than a spectral library match without RT matching. This is done because this lowers false positive molecular species level annotations and prefers species level annotations, if diagnostic fragments of multiple molecular species are present.
We want to point out that we neither prefer or disregard the MSI or Exposomics scale, but believe the more granular approach of the Exposomics scale allows a more intuitive user experience when sorting the feature table by annotation confidence.
Ranking according to MSI scale¶
See: https://link.springer.com/article/10.1007/s11306-007-0082-2
Level 4 - Unknown compounds: Not annotated, m/z-only compound database annotation (no RT, RI, CCS)
Level 3 - Putatively characterised compound classes: Compound database annotation backed by Sirius CSI:FingerId, Lipid annotation based on MS1 data
Level 2 - Putatively annotated compounds: Spectral library match to reference libraries, Lipid annotations with MS2 information, Compound database matches with m/z and RT or RI information.
Level 1 - Identified compounds: At least two orthogonal metrics confirmed with standards in the same laboratory. This annotation level is not achieved in mzmine, as it would require differentiation between lab-internal and lab-external RT and RI measurements.
Ranking according to Exposomics scale scale¶
See: https://pubs.acs.org/doi/10.1021/es5002105, https://pubs.acs.org/doi/10.1021/acs.est.0c05713
Level 5 - Exact mass of interest: Not annotated, compound database annotation (m/z only, without isotope matching), MS1 only lipid annotation.
Level 4 - Unequivocal molecular formula: Compound database match with isotope score >= 0.75
Level 3 - Tentative candidates: Compound database match confirmed with Sirius CSI:FingerId
Level 2b - by diagnostic fragments: MS2-based species level lipid annotation
Level 2a - by a library spectrum match: Spectral library match to a reference library, molecular species level lipid annotation.
Level 1 - Confirmed structure: Spectral library match to a reference library including RT or RI matching.
Annotation quality summary¶
The annotation quality summary (AQS) is a universal quality summary available for spectral and compound database matches and lipid annotations (since mzmine 4.9).
The AQS relies on spectral match scores (MS2), isotope pattern fit (IP) and deviation of m/z, RT, retention index (RI), and CCS library entries. These summaries may be disabled by default, e.g. no IP score will be displayed for GC-EI workflows, as will RI for LC-MS.
An example is shown below:
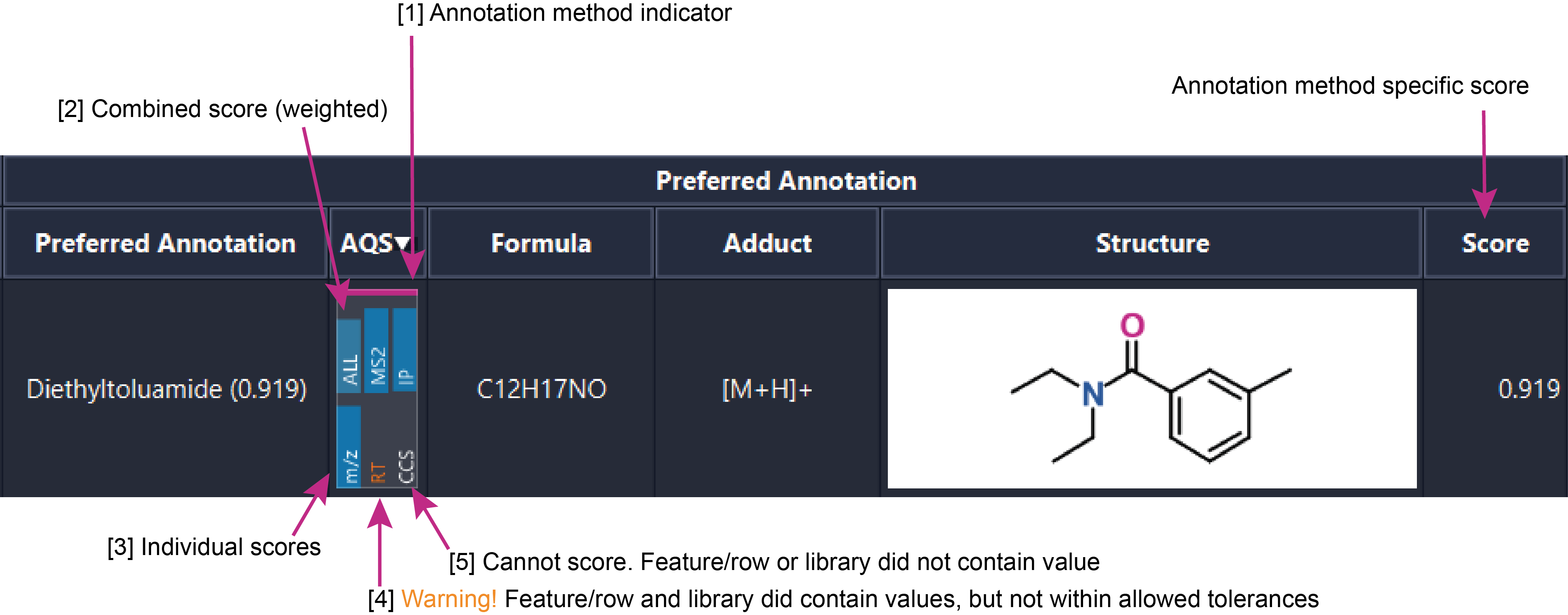
- The colored bar will indicate the annotation method this match originates from. Spectral library, Compound database, lipid annotation
- A combined score - weighted according to the annotation ranking module. See Preferred annotation.
- The individual scores, the larger the bar the better.
- The orange text indicates a warning, not an error. The library entry did contain a value for the given measurement, but it did not match. E.g. the library creator's HPLC method did not match your HPLC method.
- No bar, white text. The library did not contain a value for the given measurement, cannot score.