Batch spectral library generation¶
Feature list methods → Export feature list → Spectral library batch generation
After matching with a compound database table, only annotated spectra are exported with this module. This enables the automatic generation of mass spectral libraries with different data format provided. Different quality parameters and how to handle chimeric spectra can be set to increase the spectral quality in the new spectral library and to flag chimeric spectra. Furthermore, different information about the acquired data can be added, for example, instrument, who acquired the data,... Single best spectra but also merged spectra are exported (same energy, multiple collision energies, or multistep fragmentation)
Parameters¶
Export file¶
The path and name of the exported library spectra.
Export format¶
Here, you can set the export format. It mostly depends on your next steps and where to import the library. Every format works, but we recommend the .json format.
MS level filter¶
If data were acquired with multistep fragmentation, the MS2 or all MSn data can be exported individually. If you only acquired MS2 data or want to export MSn, the setting can remain MSn
Metadata¶
Various information about the acquired data can be stored her. For example, who acquired the data, which instrument was used, ect. On the description information about the compounds or library can be given. The Ion mode is only need, when the polarity is not safed in the data files.
Merge & select fragment scans¶
This parameter controls how fragment spectra are filtered, merged, and selected for downstream analysis (see detailed description). Briefly, either choose preset based spectral merging, input scans without merging, or an advanced setup for more options.
For library generation, mzmine recommends the simple preset Representative scans or MSn tree.
Handle chimeric spectra¶
This option checks the MS1 scans for chimeric precursor selection. A minimum for this purity can be
set and how to handle chimeric spectra.
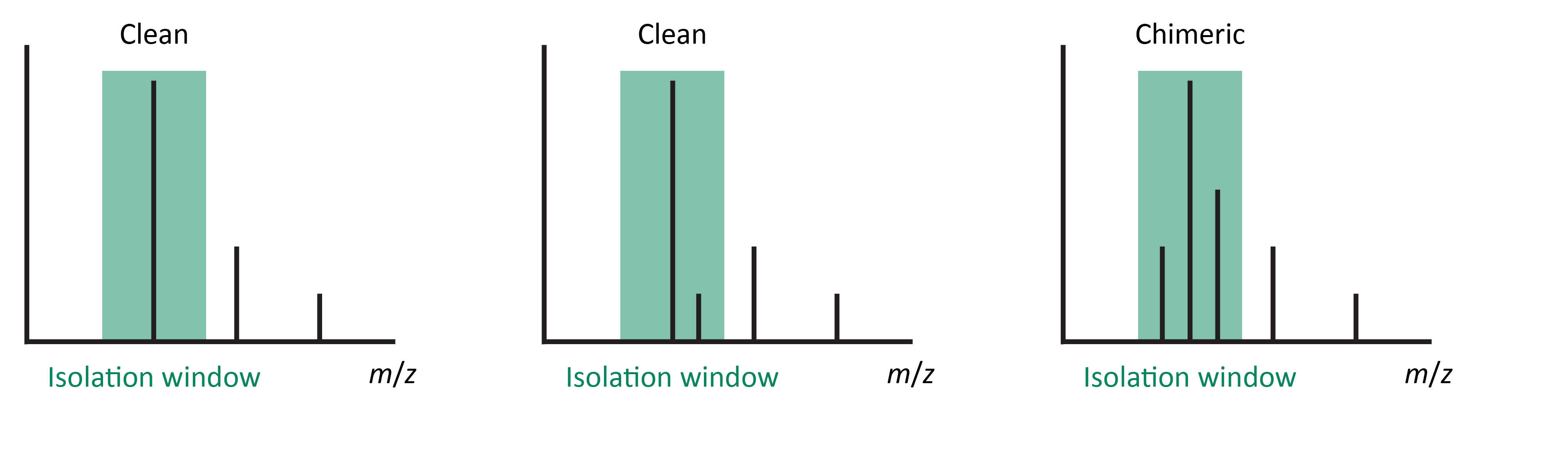
Target ion m/z tolerance¶
This is the mass tolerance for the precursor signal. The tolerance is usually smaller than the precursor isolation window and depends on the accuracy and resolution of the instrument.
Precursor m/z isolation tolerance¶
This should be the set precursor isolation window, to check for co-isolated signals in this mass window. Be aware, that some QTOF instruments have a wider isolation window than a unit, resulting in co-isolation of isotope signals.
Minimum precursor purity¶
MS1 spectra with a precursor signal explaining less then X% of the total signal intensity within the set precursor isolation window will be flagged as chimeric.
Chimeric spectra¶
Here, spectra that are flagged as chimeric can be skipped or flagged. We suggest that flagging is a better option, as the flagging state will be exported and can be removed afterwards as it contains still information about the precursor ion. But matches need to be inspected carefully.
Quality Parameters¶
Quality parameter can be set before exporting the spectra. Only spectra that fullfil those criteria will be exported.
Minimum number of signals¶
Number of signals an MSn scan must contain to be exported. Depends also on the fragmentation pattern of the compounds (very rich pattern, more signals, less informative spectra less signals). At least 3 signals might be a good compromise if the compounds show a very different fragmentation behavior.
Minimum explained signals (%)¶
The number of explained signals within the MSn scan based on a subformula level, without taking the chemical structure into account.
Minimum explained intensity (%)¶
The minimum number of the explained spectra intensity. This increases the value of more abundant signals that should be explained in a fragmentation spectrum based on the subformula level for an annotated compound. Low abundant signals can correspond to unfiltered noise, ect. and shouldn't be taken into account in the same way as high abundant signals. Therefore, we recommend this setting before the explained signal one.
Formula m/z tolerance¶
This is the m/z tolerance to assign MSn signals to a subformula.