MS2 Scan Pairing¶
Description¶
Feature list methods → Processing → Assign MS2 to feature
This module allows to pair MS2 scans with features. It assigns all MS2 scans within range to all features in chosen feature list.
Parameters¶
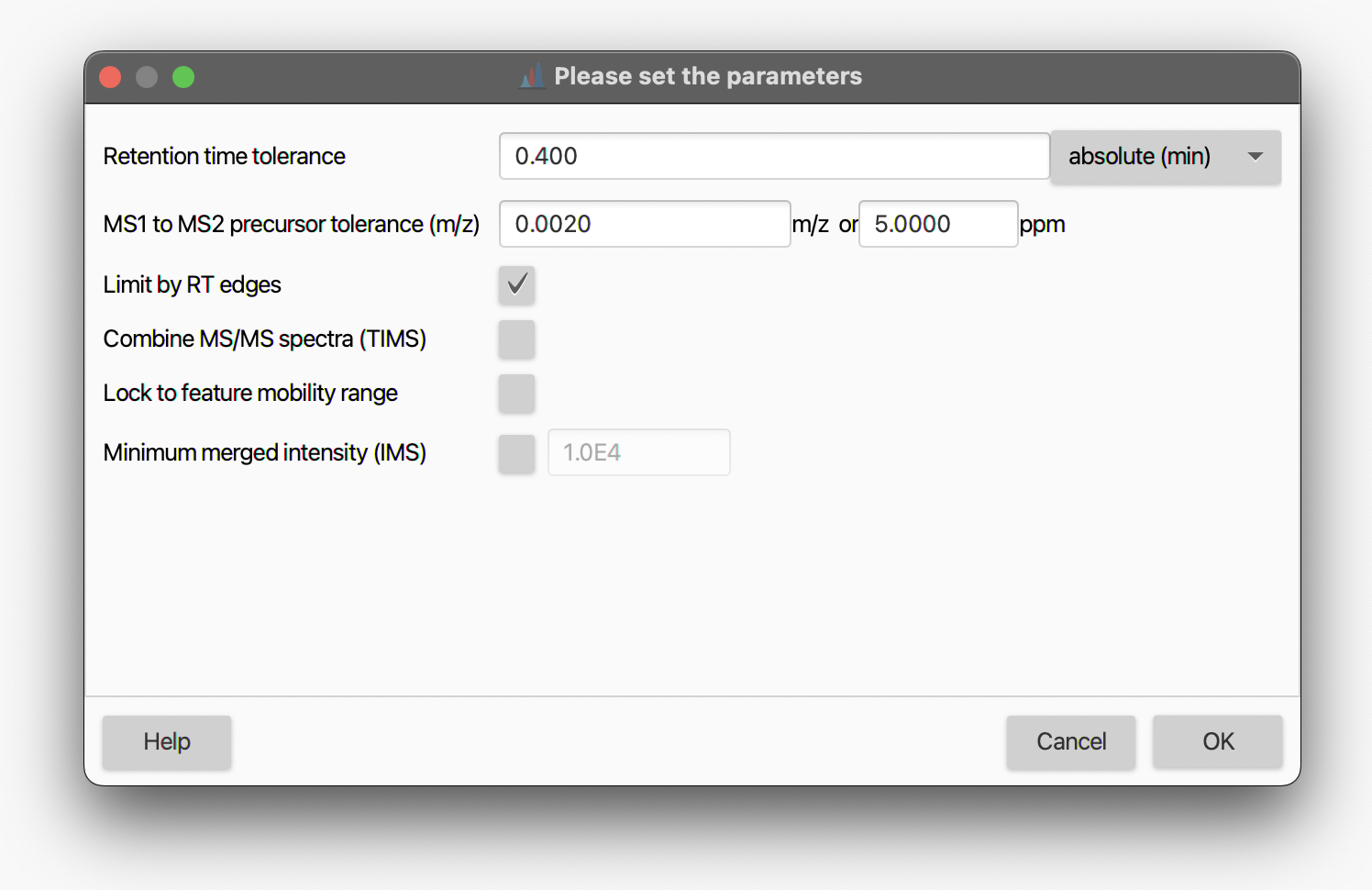
Retention time tolerance¶
The maximum offset between the highest point of the chromatographic peak and the time the MS2 was acquired.
MS1 to MS2 precursor tolerance (m/z)¶
Describes the tolerance between the precursor ion in a MS1 scan and the precursor m/z assigned to the MS2 scan.
Limit by RT edges¶
Use the feature's edges (retention time) as a filter.
Combine MS/MS spectra (TIMS)¶
If checked, all MS/MS spectra assigned to a feature will be merged into a single spectrum.
Lock to feature mobility range¶
If checked, only mobility scans from the mobility range of the feature will be merged.

Minimum merged intensity¶
If an ion mobility spectrometry (IMS) feature is processed, this parameter can be used to filter low abundant peaks in the MS/MS spectrum, since multiple MS/MS mobility scans need to be merged together.