Ion identity networking¶
Feature list methods → Feature grouping → Ion identity networking
Annotates grouped features (same retention time + optionally, feature shape and height correlation) as ion adducts, in source fragments, and multimers. Also used as input to the [Ion Identity Molecular Networking workflow] (https://ccms-ucsd.github.io/GNPSDocumentation/fbmn-iin/) on GNPS. Searches all feature pairs against an ion library for each possible combination. Great in combination with the Molecular Networking module that calculates fragmentation pattern similarities in mzmine and allows for interactive visualization.
Warning
Apply metaCorrelate before running this module
Recommended citations¶
Info
When using this modules, please consider citing the corresponding publication(s):
Schmid, R., Petras, D., Nothias, LF. et al. Ion identity molecular networking for mass
spectrometry-based metabolomics in the GNPS environment. Nat Commun 12, 3832 (
2021). https://doi.org/10.1038/s41467-021-23953-9
Schmid R., Heuckeroth S., Korf A., et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3, In Review (2023)
Parameters¶
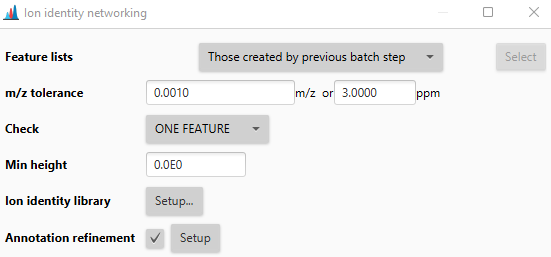
m/z tolerance¶
Intra sample m/z tolerance describes the difference between two ions of the same molecule at the same retention time. This tolerance is usually very small and depends on the mass resolution. Orbitrap instruments for example 3 ppm.
Check¶
- ONE FEATURE: Only one feature needs to match (m/z difference within one sample)
- ALL FEATURES: All features need to match (m/z difference within ALL samples)
- AVERAGE: Matches the m/z difference of average values
Min height¶
Minimum height of features to consider. Leave at 0 to use all features that passed the feature detection workflow criteria.
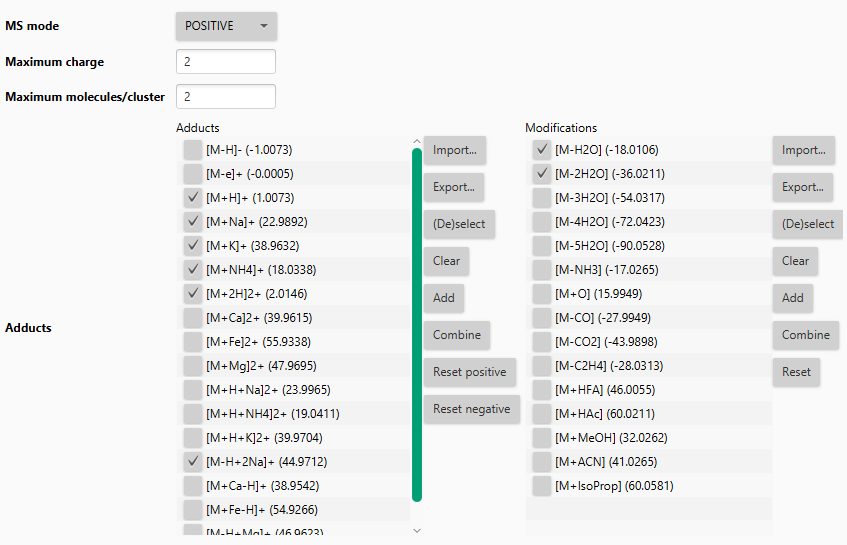
Ion identity library¶
Defines the ions to search. When two features, applied with two ions, result in the same neutral mass - an annotation is made. New ions can be defined. Adducts (left) and neutral modifications (right) are combined to create the final ion library.
Tip
This step should only focus on the main ions that are typically detected in the MS method. Later the Add more ion identities module can add strange ions to existing networks.
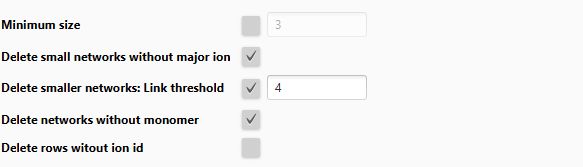
Annotation refinement (optional)¶
Annotation refinement is optional but should be applied to finalize ion identities, after all subsequent optional steps of Add more ion identities steps.
Parameters:
- Minimum size (optional): Only retain ion networks with at least n ions (often 3). The more ions the higher the confidence in the annotation.
- Delete small networks without major ion (optional): Major ions are defined as M+H, M+Na, M+NH4, M-H2O+H
- Delete smaller networks: Link threshold (optional): Important parameter to only keep the best annotation, when this annotation is supported by n-1 ions (network size n).
- Delete networks without monomer (optional): Only keep a network if at least one is M+... and not a multimer like 2M, 3M...
- Delete rows without ion id: Remove all rows from the feature list that have no ion identity annotation