Retention time correction of scans¶
Note
This module is currently in beta phase.
Corrects retention time drifts by searching for standards across selected samples and corrects retention time drifts between samples by changing the scan retention times.
Implications and restrictions¶
Correcting retention times of scans instead of features has certain implications and up- and downsides.
Pros:¶
- All visualizations in mzmine will use the calibrated retention times to visualize.
- this means that e.g. the Raw data overview or the feature table will visualize corrected chromatograms
- Gap filling will use the corrected retention times, which may lead to less misalignments during gap filling of complex samples
Cons:¶
- Corrected scan retention times must still be in strongly monotonous order (cannot swap scans)
- This means that large RT shifts cannot be mitigated
- Irregular shifts cannot be corrected: If feature A at RT 5.4 is shifted to higher RT and feature B
at RT 5.4 is shifted to lower RT the effects may cancel each other out
- Such shifts may happen if the stationary phase is damaged/overloaded or the pH of the eluent changes and thus affects compounds with different pKs differently
- All files must start at 0.0 min, no negative RTs possible.
Tip
The retention time correction may be applied multiple times to correct larger shifts.
Exemplary results¶
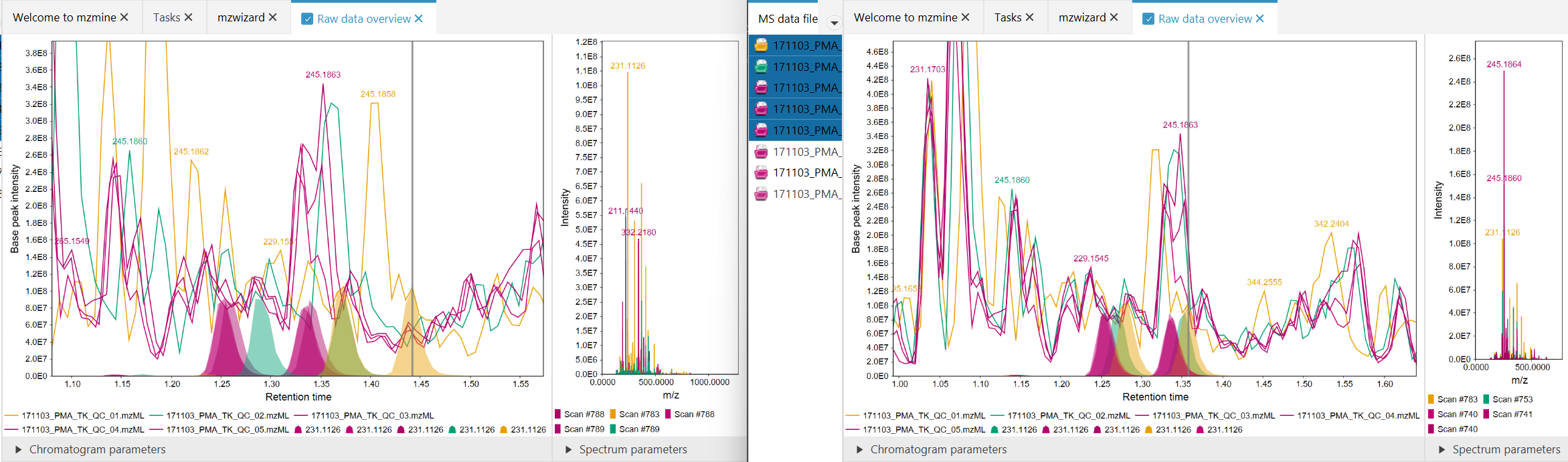 Figure 1: Before (left), After (right). Significantly better alignment was achieved for the orange
data file.
Figure 1: Before (left), After (right). Significantly better alignment was achieved for the orange
data file.
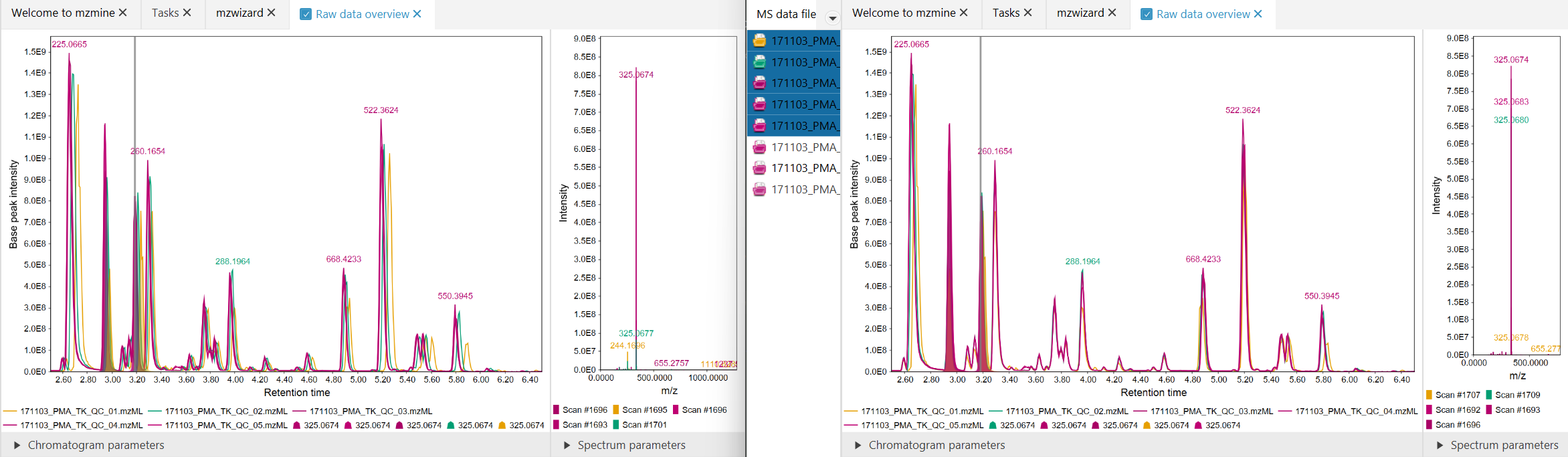 Figure 2: Before (left), After (right). Significantly better alignment was achieved for the orange
data file.
Figure 2: Before (left), After (right). Significantly better alignment was achieved for the orange
data file.
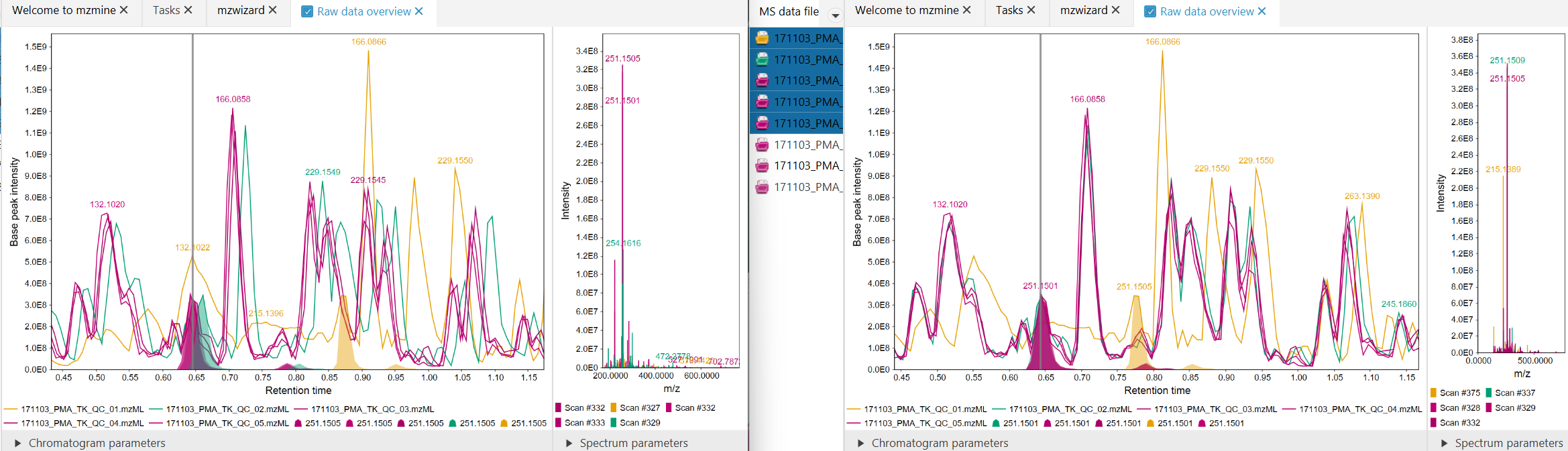 Figure 3: Before (left), After (right). Large retention time shift of the orange signal. After
correction, the signals move closer together, but the large shift cannot be corrected completely due
to the mentioned restrictions.
Figure 3: Before (left), After (right). Large retention time shift of the orange signal. After
correction, the signals move closer together, but the large shift cannot be corrected completely due
to the mentioned restrictions.
Standard selection¶
The calibration module will look for features that appear in all feature lists that are selected as reference samples. Ambiguity will be avoided during standard selection by also examining the same mz tolerance but three times the RT tolerance window around a potential standard. If the extended window contains more matching features in any sample, a high MS2 similarity is required or the feature will not be used as a standard.
Parameters¶
Feature lists¶
The feature lists whose data files shall be corrected. Each feature list may only contain a single raw data file. Visualizers will need to be reopened to show the result.
It is recommended to use resolved feature lists for this step. Since the scans are corrected, all other (e.g. prior) feature lists using the same data file will be affected as well, once a new processing step is applied (RTs are not recalculated automatically for old feature lists).
Reference samples¶
Select the sample types (from the metadata table) that shall be used as reference to search standards. The created calibration will be interpolated onto the other data files by weighting the nearest two standard files by their acquisition date.
mz tolerance¶
Maximum mz deviation for potential standards.
Retention time tolerance¶
Maximum RT tolerance for potential standards.
Minimum standard intensity¶
The minimum required intensity of a feature to be considered a standard for RT calibration.
RT standard calculation¶
Select if the corrected standard value to which the samples shall be calibrated to shall be chosen as average or median.
Correction method¶
Select the method which shall be used for correction.
Multi linear correction¶
- Calculate difference of standard and specific RT in a sample.
- Smooth difference using a moving average of num_standards * bandwidth points.
- Apply asymmetric least squares regression (ALS) on the smoothed differences
- Apply loess interpolation on current retention times (domain values) vs the ALS result (range values)
- calculate potential new differences (= correction values) from the loess fit
- ensure monotonicity of the new fit
- create a final loess fit of the corrected differences
- apply correction to original scan RTs
Interpolation bandwith¶
Define a percentage of standards for moving average calculation.