Additional tools
Chromatogram plot¶
Description¶
Visualization → Chromatogram plot
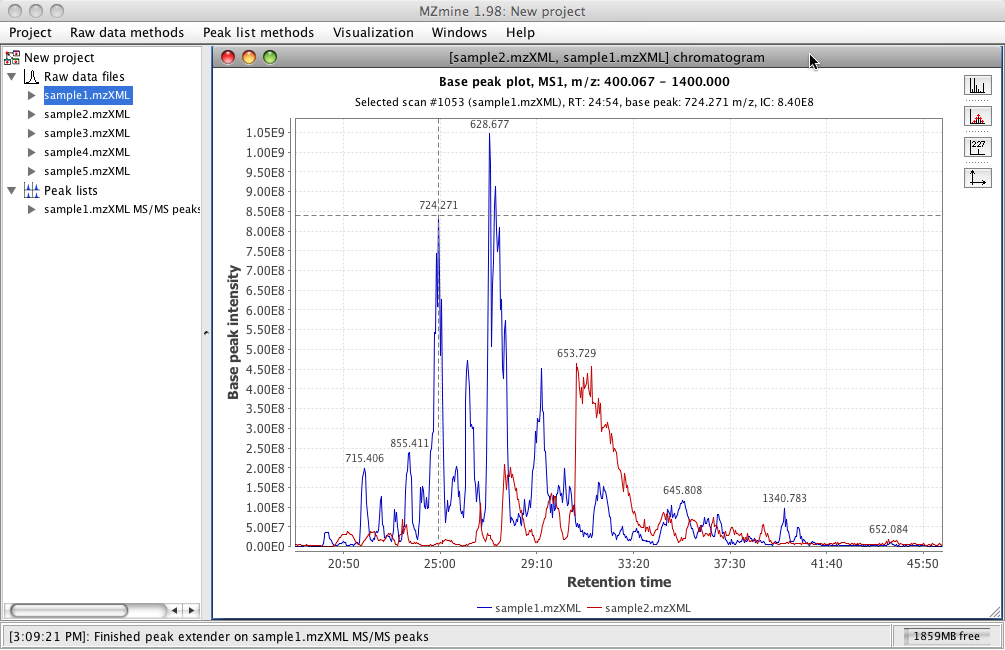
There is a possibility to display chromatographic peaks outside of raw data overview. This standalone two-dimensional plot visualizes TIC/XIC data. All the chromatograms are displayed in the same plot.
The x-axis corresponds to retention time and the y-axis is the intensity level of the signal.
Parameters¶
Raw data files¶
List of raw data files to display in the TIC visualizer.
MS level¶
Scan level (MS1,MS2,... ,MSn) to display in the plot.
Plot type¶
TIC or base peak
Retention time¶
Retention time (x-axis) range.
m/z range¶
Range of m/z values. If this range does not include the entire scan m/z range, the resulting visualizer is XIC type.
Selected peaks¶
List of chromatographic peaks to display in the TIC visualizer. This option is available only if a peak list related to the selected raw data file exists in the current project.
MS spectrum¶
Visualization → MS spectrum
Displays all the ions from a selected scan. Can be used to explore mass spectrum outside of raw data overview. Only one raw file can be chosen.
It shows a plot of two dimensions, where X axis corresponds to m/z value and Y axis is the intensity of the ion signal.
Parameters¶
Scan number¶
Choose the scan to visualize
2D visualizer¶
Visualization → 2D plot
This tool displays a plot of two dimensions, where X axis corresponds to retention time and Y axis is the m/z value. This visualization of spots in the plot corresponds with the intensity of the data in that region.
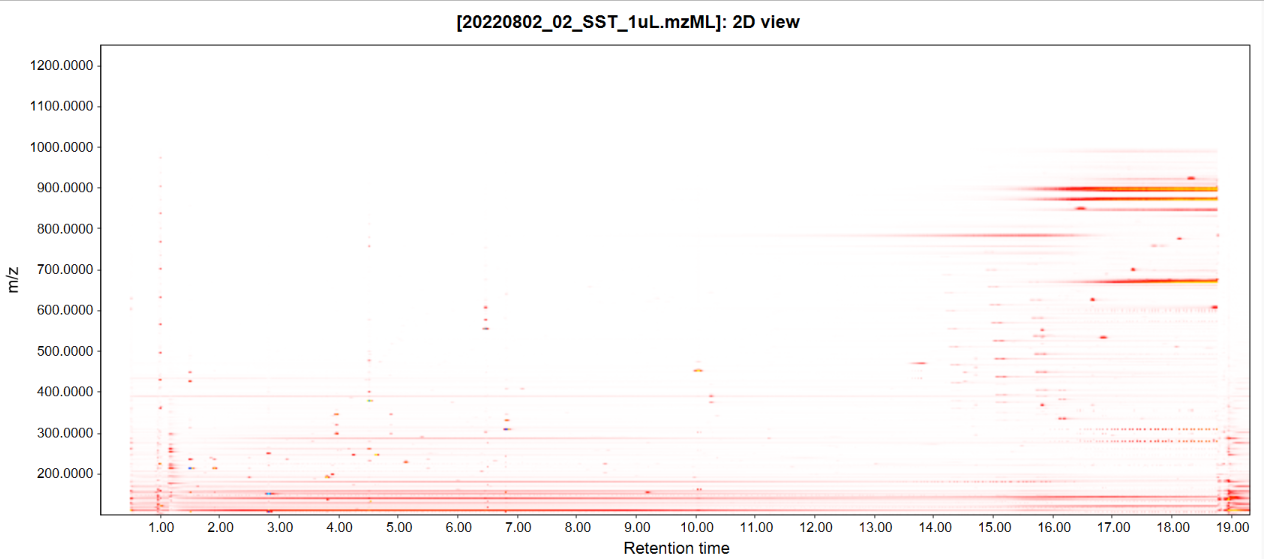
User can define features from the feature list to be displayed on the plot.
Parameters¶
Type of data¶
This plot can use either resampled data as input (faster), or raw data (slower).
Scans¶
Here user can set up the level (MS1,MS2,... ,MSn), polarity, retention time, and the other parameters of the scans to be used.
m/z¶
Defines range of m/z values.
3D visualizer¶
Description¶
Visualization → 3D plot
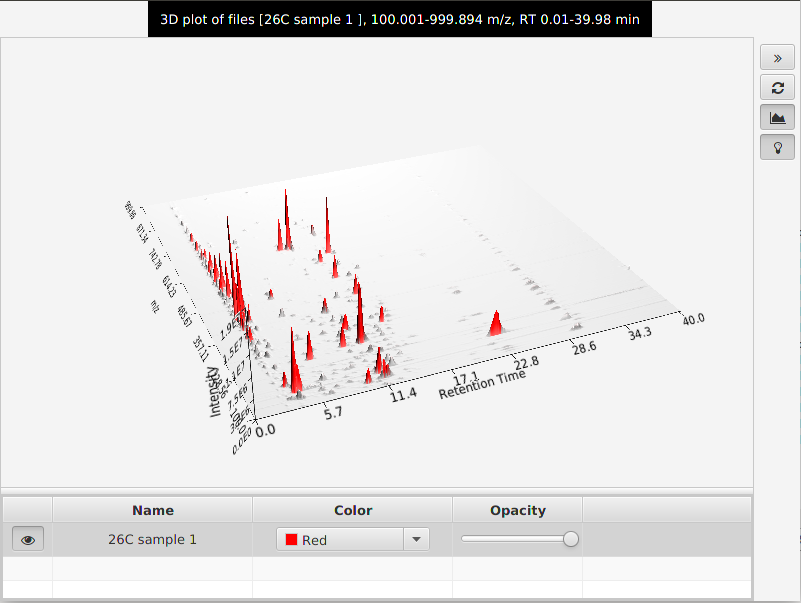
This tool presents a three dimensional plot where X axis represents the retention time, Y axis the m/z value and Z axis the intensity of the signal. This plot is the collection of all the information from the raw data in a graphical representation.
Parameters¶
Scans¶
Here user can set up the level (MS1,MS2,... ,MSn), polarity, retention time, and the other parameters of the scans to be used.
m/z range¶
Range of m/z values.
Features¶
Features that will be taken as an input.
Retention time resolution¶
Number of data points on retention time axis.
m/z resolution¶
Number of data points on m/z axis.
Scan histogram¶
Description¶
Visualization → Scan histogram
This module creates m/z distribution histograms of all m/z values in mass lists across specified scans. The binning width, in which the m/z values are counted, can be changed dynamically. The number of scans that contain a specific m/z value (bin) are plotted.
Parameters¶
Scans¶
Here user can set up the level (MS1,MS2,... ,MSn), polarity, retention time, and the other parameters of the scans to be used.
m/z¶
Limit the range of the histogram (can improve performance).
Signal intensity range¶
Optional parameter
Allows to limit signal intensities (can improve performance).
Mass defect¶
Optional parameter
Filters for mass defects in the signals.
Type¶
Type of the histogram to be created. Available options:
- m/z,
- Intensity,
- Intensity (noise recalibrated),
- Mass defect.
Bin width¶
The binning width to count m/z value occurence in scans.
Use mobility scans¶
If the input data has ion mobility dimension, this data can be used instead of the data from the summed frames.
Scan inject time analysis¶
MS(n) spectra tree¶