Batch mode processing¶
Besides the interactive GUI, mzmine allows the user to run processing workflows in an automated manner using the "batch mode". Entire processing pipelines (including data import/export) can be run with few clicks, or even through the command-line application. This makes MZmine suitable to be integrated into automated data analysis pipelines (e.g., QC systems).
Batch files (XML format) are essentially lists of tasks run by mzmine one after another. Any of the methods available in mzmine can be included in the batch file.
How to run batch processing¶
Project Batch mode
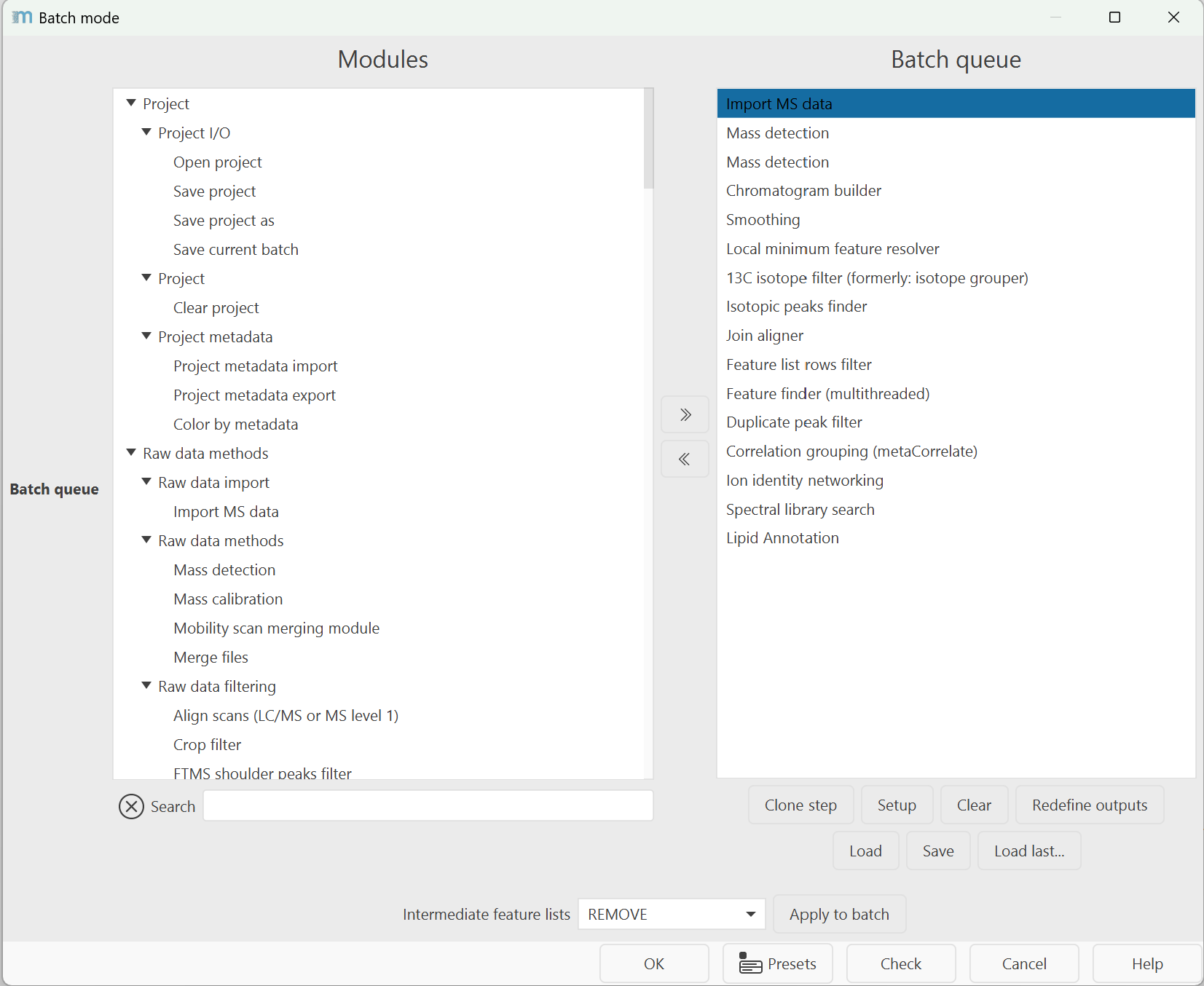
When a new step is added to the queue its parameter setup dialog is shown. The "Set parameters" button allows the user to modify a step's parameter settings. The "Clear" button removes all steps. The "Load" and "Save" buttons make it possible to read and write batch steps to XML files.
Tip
Redefine outputs is a quick aid to set a new file path and base filename to all export modules (e.g., the feature list CSV export). Each step will add a suffix for the user to identify the output files. If one module exists multiple times, the suffix will also contain a numbering.
The first step of a batch queue is performed on those raw data files and/or feature lists selected by the user. The remaining steps are performed on the results produced by each preceding step (File/Feature list selection must be set to Those created by previous batch step). For example, if the first step of the batch queue is the Chromatogram builder, it will produce feature lists as a result. If the following step were Peak list deconvolution then it will be performed on the peak lists produced by the preceding Chromatogram builder step.
Tip mzmine "remembers" the last settings used.
Tip
Intermediate feature lists is an option to apply the same handling of intermediate results to all batch steps. For example set all to keep during batch optimization or to remove/process in place, to run a final batch with maximum performance.
mzwizard¶
The mzwizard facilitates quick set up of general workflows for various sample introduction systems, (ion mobility) mass spectrometers, and workflows. This is the recommended way to configure workflows and create batch files. Batch files can then be modified further to improve results or to add more export steps.
mzwizard Processing wizard
